
After the PmiRDB project initialized, much discussion was taken to design the basic frame of this unique database. In brief, it includes three main parts 1) what kind of data will be included; 2) How to present these mega data; 3) How the database can benefit users/researchers.
In terms of one month discussion, we came to the frame as follows.
Considering the unique feature of pre-miRNAs, stem-loop structure, we would focus on the species whose genome reference sequences are available. At the same time, NGS (next generation sequencing) method greatly speeded up the research/annotation of miRNAs. sRNA-Seq datasets are another requirement. In addition, PARE-Seq (Parallel Analysis of RNA Ends) method was confirmed a valuable one to validate miRNA targets. Thus, PARE-Seq datasets and subsequent analyses would be included. The evolution of miRNA is a topic where many issues are not clear. Synteny gene (miRNA gene) analysis is a powerful tool to illustrate the evolutionary processes such as gain, loss, duplication, etc. Therefore, we planned to include miRNA synteny analyses.
Genome references, sRNA-Seq datasets, PARE-Seq datasets, and Synteny analyese, etc., all together compose a huge pool of mega data. How to present these data is the key. We designed three ports to resolve this issue. First, a hierarchical system was employed to display information on miRNAs. For instance, the first level focused on species, and the second layer is a page to present all miRNA loci (MIRs, could be considered as miRNA genes even though some loci are clustered together), and the third layer is to display all details on a specific miRNA including all sequences, structure, coordinate information, targets, expression, etc. Second, to users, how to fetch/download the data they are interested. We designed an easy access by which users could download all data displayed/stored in webpages. Meanwhile, accesses for bulk and customized download ports will be deployed. To improve the accessibility of all data, we will design many occasional download links in webpages. For instance, in miRNA detail information page, users could easily download its sequences, structure, etc., or users could select miRNAs of their interest and download them in miRNA list page.
The most important part is how the database could benefit its users/researchers, which inherently require all data in PmiRDB are solid and with high quality. To meet this goal, as described previously (logs in PmiRDB project initialized, 02/01/2018), we will develop a suite of uniform tools to annotate/identify miRNAs and carry out subsequent analyses. Considering the inaccuracy and noise existed in current miRNA databases such as miRBase, we will curate all data inherited from these databases and make sure brand new data are highly confident.
The following is an overall design of PmiRDB by software XMind. We appreciated all the effort made by the former member, Dong Wang (now he is working in Alibaba), in Dr. Yang’s lab.
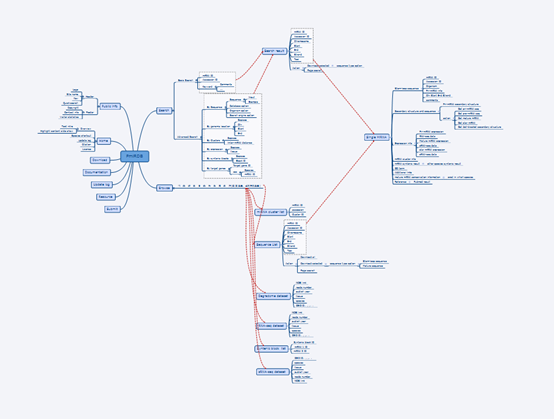
Overall design of PmiRDB
The figure below zooms in the part where a single miRNA is located and shows all information we plan to store and present in PmiRDB.
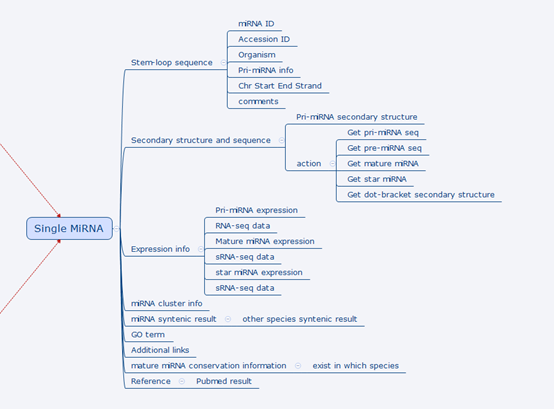
Details on a single miRNA design